IoT-based Sustainable Construction Management and Energy Lab
Dysregulation of proteolysis and proteostasis accumulates aberrant and intractable protein species in cells and often cause devastating human diseases. The central focus of our group is to understand critical proteolytic machinery, such as proteasome and deubiquitinating enzymes (DUBs), in human health and define them as promising drug targets. A hallmark of our research highlights the significance of cellular proteome balance in normal and diseased states. We will strive to pioneer the protein degradation mechanisms from both biology and therapeutic perspectives, and eventually provide new and foundational paradigms in the field.
Current
interests in the
lab can
be summarized
into a few
of subjects:
Another area of our interest is how to develop novel chemical strategies to dispose of the challenging or “undruggables” targets by induced protein degradation.
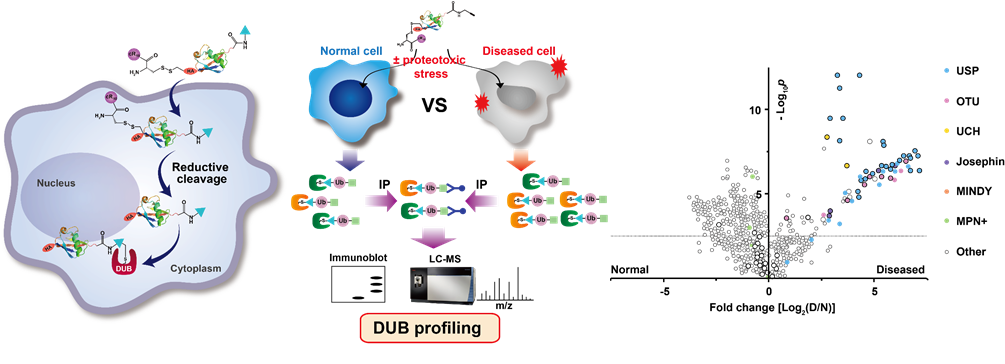
Activity-based protein profiling is a powerful method in assessing biological activities of a variety of enzymes in cell and tissue samples. Notably, activity-based DUB probes of ubiquitin (Ub)-derivatives have allowed profiling of novel DUB activities and facilitated the development of DUB inhibitors. We have been developing diverse activity-based Ub probes for in vitro and endogenous DUB labeling in normal cells and diseased cells. DUBs that are associated in the diseased states will be specifically identified from this approach.
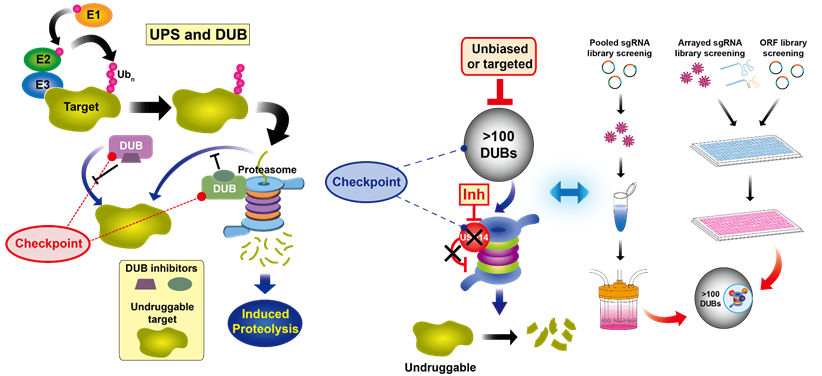
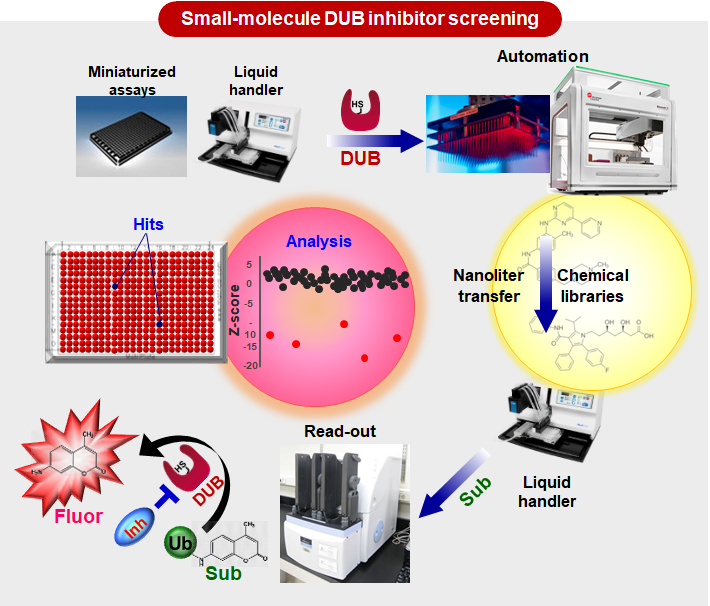
Our group is particularly interested in developing the strategies for improving proteostasis by manipulating the ubiquitin-proteasome system (UPS). Notably, DUBs may represent promising drug targets in impaired proteostasis, due to their capability of regulating protein half-lives. Therefore, DUBs that are associated with human diseases will be also identified from chemo-genomic screening approaches and then will be evaluated for their potential to regulate the stability of disease-related proteins and undruggable targets.
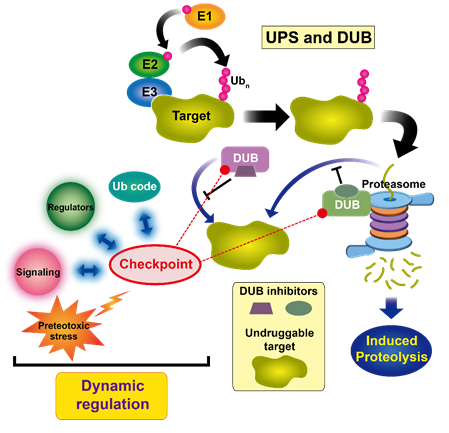
Proteasome serves as a critical hub as well as a rate limiting step for the proteolytic degradation pathway, and whose activity can be under dynamic regulation by proteasome-associated factors, such as DUBs. Thus, we will try to investigate dynamic nature of DUB reactions on the proteasome and upstream of the proteasome for offering fundamental basis of proteolytic versatility.
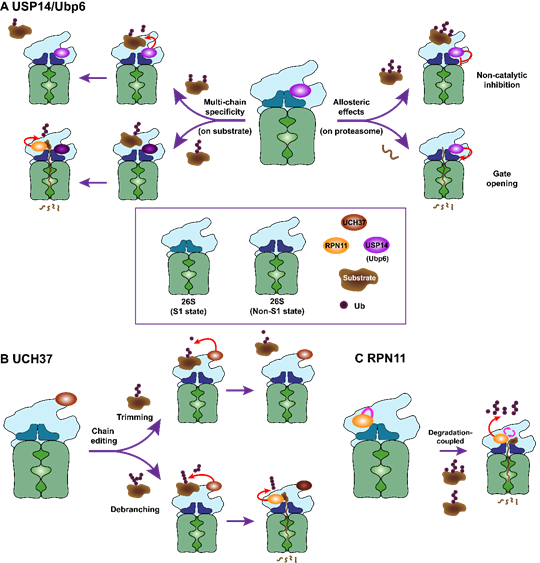
Given that DUBs are critical checkpoints in the proteolytic pathway and proteasome is the rate limiting step for substrate engagement in protein degradation, we try to develop novel class of chemical proteolytic inducers for directly engaging DUBs or proteasome into the undruggable targets. These DUB- or proteasome-guided degradation inducers may significantly innovate the conventional paradigm of targeted protein degradation because they might bypass the obligatory requirement of polyubiquitin chains as the typical degradation signal.
본 사이트는
Internet Explorer 8 이하 버전을
지원하지 않습니다.
Internet Explorer 9 이상으로 업데이트 하거나
크롬, 파이어폭스, 오페라, 사파리 최신 브라우저를 이용해 주십시오.
불편을 드려 죄송합니다.